

A online service supports the transformation and the registration of your own tractography bundle from the trk format in individual space to ply format in the specimen space.
A repository of python scripts useful to manipulate the
3D models, the annotations and the bundles as point
clouds or meshes. Try out to clone the repository here
https://github.com/minilabus/bradiphopy
SERVICE AND SOFTWARE
The result of the transformation and registration of the uploaded bundles available for download.
The
PLY format is the best option for the visualization with CloudCompare tool. The VTK format is encoding the
streamlines only as polylines.
The bundle tractography can be in TRK or TCK format (no whole brain tractogram, size limit is 200MB). Only a single bundle can be specified at each time. The reference image has to be a skull-free FA map as nifti file. This file is mandatory and refers to the reference space of the bundle tractography. The particular template should be provided to verify the compliance of a standard space.
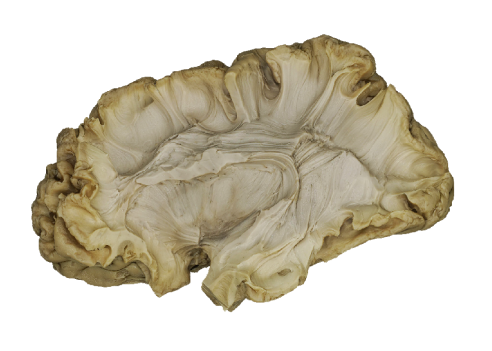

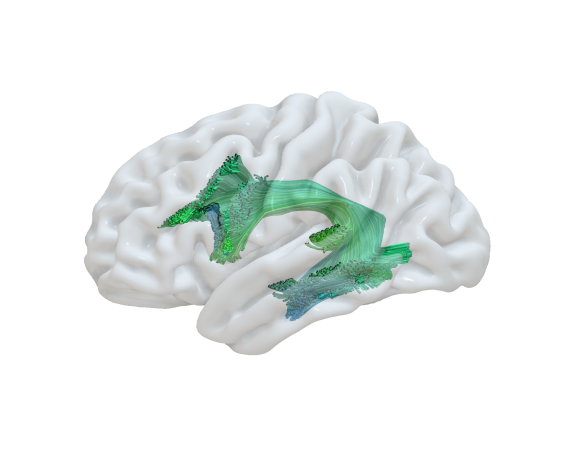
Every bundle tractography obtained from DWI can be visualized as overlay with the 3D models of ex-vivo dissection. The projection of a bundle tractography is carried out by a step of registration and a step of conversion of streamlines as 3D tube. A nonlinear Registration Step is performed to the reference space of the specimen. By uploading a pair of files, a TRK (or TCK), and its matching skull-less FA map as Nifti file, respectively, you may obtain the correspondent representation suitable for a concurrent visualization with 3D dissection models. The uploaded files should be anonymized and they will be deleted after the computation.
