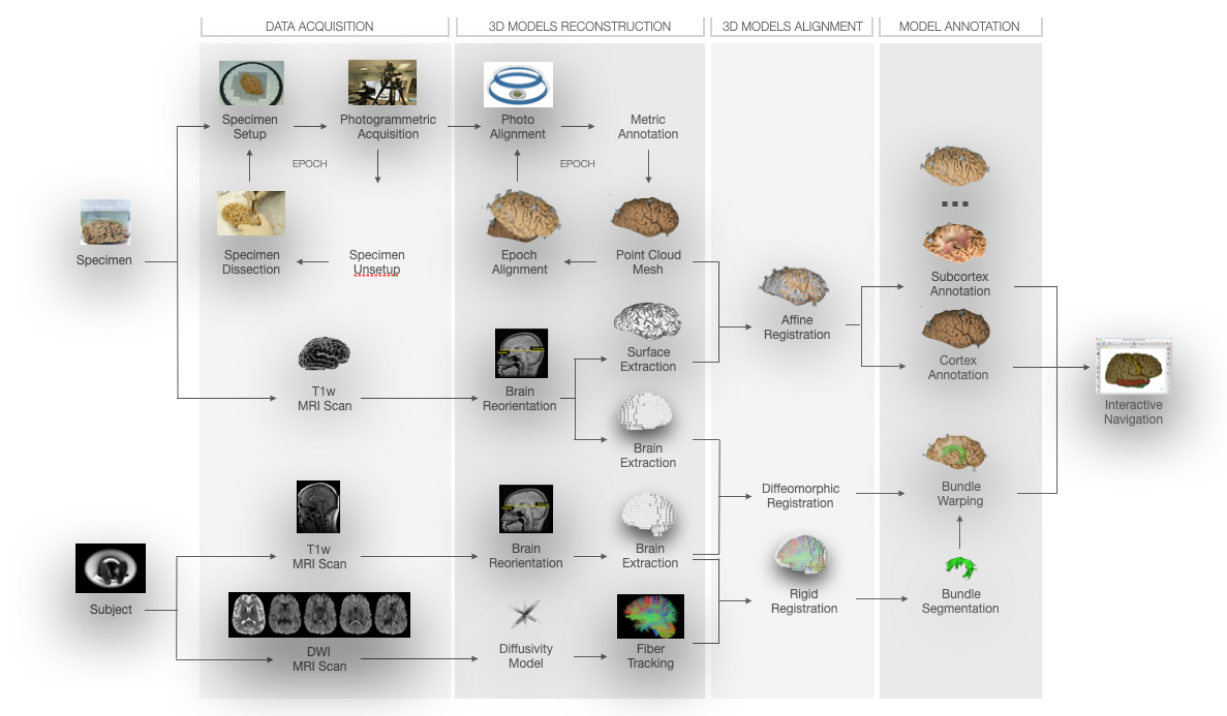
The photogrammetric ex-vivo models, their cortical and subcortical annotations and the tractography bundles are available for download as open data, both as point clouds and meshes.


A online service will convert your bundle tractography in a format compliant with the 3D Web Viewer. A repository of python script will enable you to manipulate your annotation of ex-vivo models.


The annotation of neuroanatomical landmarks, both cortical and subcortical, on a 3D photogrammetric model
reconstruction of ex-vivo specimen, can be translated to an in-vivo template of the brain using magnetic
resonance.
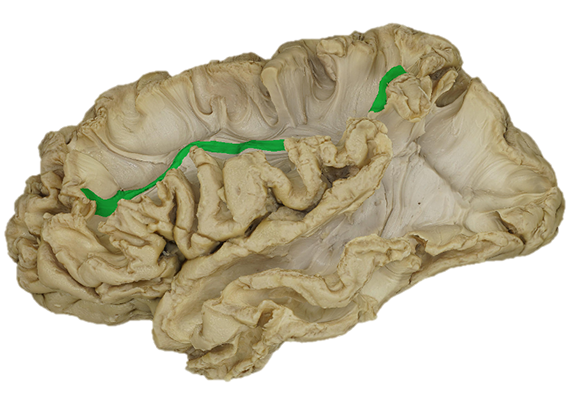
A segmentation of white matter ROIs based on ex-vivo dissection can be available in the
standard MNI space.
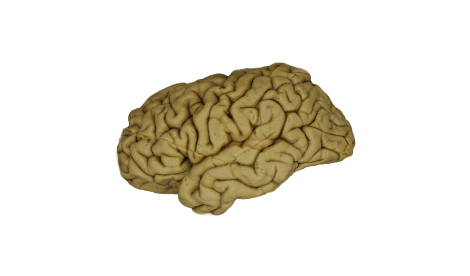
EX VIVO | IN VIVO
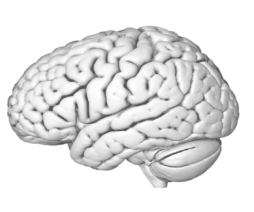
IN VIVO | EX VIVO
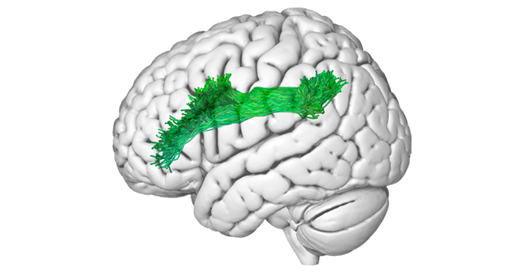
The in-vivo detection of the main white matter structures using tractography with DWI can be translated to an ex-vivo 3D model of a specimen reconstructed using photogrammetry. The pathways of fibers encoded in a MNI reference space can be superimposed on the texture of the subcortical tissue in the ex-vivo specimen space.
3D visualisation with BraDiPho

Merging diffusion tractography with photogrammetric dissection of the human brain. User-friendly framework to explore microdissection and tractography data within the same environment.
BRADIPHO
Fiber Bundles